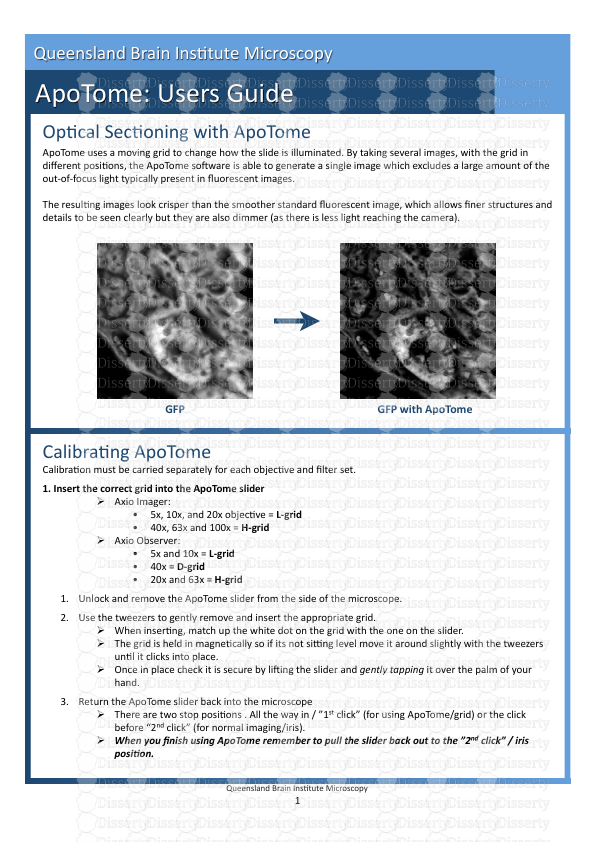
Op#cal Sec#oning with ApoTome ApoTome uses a moving grid to change how the slid
Op#cal Sec#oning with ApoTome ApoTome uses a moving grid to change how the slide is illuminated. By taking several images, with the grid in different posi#ons, the ApoTome so?ware is able to generate a single image which excludes a large amount of the out‐of‐focus light typically present in fluorescent images. The resul#ng images look crisper than the smoother standard fluorescent image, which allows finer structures and details to be seen clearly but they are also dimmer (as there is less light reaching the camera). Calibra#ng ApoTome Calibra#on must be carried separately for each objec#ve and filter set. 1. Insert the correct grid into the ApoTome slider Axio Imager: • 5x, 10x, and 20x objec#ve = L‐grid • 40x, 63x and 100x = H‐grid Axio Observer: • 5x and 10x = L‐grid • 40x = D‐grid • 20x and 63x = H‐grid 1. Unlock and remove the ApoTome slider from the side of the microscope. 2. Use the tweezers to gently remove and insert the appropriate grid. When inser#ng, match up the white dot on the grid with the one on the slider. The grid is held in magne#cally so if its not siXng level move it around slightly with the tweezers un#l it clicks into place. Once in place check it is secure by li?ing the slider and gently tapping it over the palm of your hand. 3. Return the ApoTome slider back into the microscope There are two stop posi#ons . All the way in / ”1st click” (for using ApoTome/grid) or the click before “2nd click” (for normal imaging/iris). When you finish using ApoTome remember to pull the slider back out to the ”2nd click” / iris posi<on. 1 Queensland Brain Ins#tute Microscopy ApoTome: Users Guide Queensland Brain Ins#tute Microscopy GFP GFP with ApoTome Calibra#ng ApoTome 2. Phase CalibraAon This calibra#on requires the mirrored calibraAon slide. 1. Focus on the cross‐hair at the centre of the silver square on mirrored calibra#on slide. Ensure you are using the BF RL (brighEield reflected light) filter set and the HXP lamp. The light shining on the slide should appear yellow‐green and down the eyepieces the cross‐hair should appear black. 2. Once you have found the cross‐hair switch to camera mode and ensure the cross‐hair is focused as best you can. If the cross‐hair is out of focus you will not calibrate ApoTome correctly. 3. In the top menu go to AcquisiAon > ApoTome > Phase CalibraAon 4. In the window that appears make sure you have the right objec#ve selected, the correct grid size selected and that you are using the BF RL filter. 5. Click Next. 6. Now adjust the exposure of the camera for imaging the cross‐hair. It should adjust automa#cally by clicking measure. If not, bring the exposure of the camera to a point where there is maximum brightness but no overexposure. 7. Click Next. 8. Push the ApoTome slider all the way into the microscope. You should be able to see grid lines across the live image. 9. Leave the exposure as it is and posi#on the green rectangle over an area containing roughly 10 grid lines. This is the area axiovision will look at to automa#cally focus on the grid lines. Make sure the rectangle is not over the cross‐hair. 10. Click Full Scan. 11. The Grid will move through a series of posi#ons, un#l it finds a posi#on where the grid lines are in focus. The graph showing the posi#ons should have a defined peak at the best loca#on. If there is no defined peak ApoTome is not calibraAng correctly. 2 Queensland Brain Ins#tute Microscopy Out of Focus Correctly Focused Calibra#ng ApoTome 12. Click “Local Scan” to ensure axiovision has found the best grid posi#on. Once this is complete click Next. 13. This step requires you to straighten the camera. Move the stage slightly so that the crosshair cannot be seen then gently twist the camera un#l you the angle reads 0.0 (or as close as possible). 14. Click Next. 15. You will now need to do another Full Scan. If ApoTome is working correctly the graph will look similar to below. Once complete click Local Scan, then click Next. 16. The phase calibra#on is now finished. If you change objec#ves you will need to calibrate for the new objec#ve. 3. Grid Focus CalibraAon This calibra#on step is to configure ApoTome for each fluorescent marker you are using. 1. Remove the mirrored calibraAon slide and focus on your sample slide. 2. Under the drop down menu go to AcquisiAon>ApoTome>Grid Focus CalibraAon. 3. The first window that appears allows you to choose which filter set you wish to calibrate for (e.g. FITC or DsRed). You do not need to change any of the other seXngs here. Select the appropriate filter and click Next. 4. Adjust the exposure by clicking Measure, then click Next 5. Push the ApoTome slider into place – the grid should be faintly visible over the fluorescence of your sample. 3 Queensland Brain Ins#tute Microscopy Calibra#ng ApoTome 6. Draw the green selec#on rectangle over a bright region of the image (this will help axiovision focus the grid). Once you have selected a bright area click Full Scan. If you can’t see the grid lines during the full scan, either the sample is too dim for ApoTome, or ApoTome is not calibraAng correctly. If it has calibrated you will see a defined peak and the grid lines will be visible on the image. 7. Once you have completed the Local Scan click Next. 8. ApoTome is now calibrated for this marker. You will need to run this calibra#on again for each fluorescent marker you are using. It is necessary to create a new calibra#on for each marker but also for each light source (i.e. separate calibra#ons will be required to excite GFP with the colibri LEDs and the external HXP lamp). Important SeXngs for ApoTome Click on ApoTome in the workarea to the le?. Under the seYngs tab select: 1. The correct grid for the objec#ve being used. 2. The “Grid Visible” op#on under Live Mode – this will ensure the fastest refresh rate for live mode 3. The “OpAcal SecAoning” op#on under Acquisi#on Mode – if not checked op#cal sec#oning will not occur 4. A weak ApoTome filter is recommended if gridlines are s#ll visible in the final imaged. This filter is used to remove any bands le? by the grids but a good calibra#on should avoid this. 5. 2x Averaging (noise reduc#on) is recommended as an ini#al seXng. You may not need this if your sample is bright, likewise may need to increase this with par#cularly dim samples. Under the extras tab ensure the display normalizaAon box is #cked. ApoTome images are very dim as most of the light is being removed ‐ Display Normalisa#on rescales how the images are displayed so you can see them properly. If you are using ApoTome but your images appear completely black this op#on is most likely disabled. Once ApoTome has been calibrated you can image as normal (e.g. using Live/Snap or MulAdimensional AcquisiAon experiments). ApoTome opAcal secAoning will occur whenever the ApoTome slider is pushed all the way into the microscope. 4 Queensland Brain Ins#tute Microscopy uploads/Geographie/ apotome-guide.pdf
Documents similaires










-
16
-
0
-
0
Licence et utilisation
Gratuit pour un usage personnel Attribution requise- Détails
- Publié le Jui 26, 2022
- Catégorie Geography / Geogra...
- Langue French
- Taille du fichier 0.7521MB